The Tans Lab
Biophysics @ AMOLF Amsterdam
About the group
We study the dynamics of single proteins and single cells in organoids using novel experimental approaches.
The group is based at the AMOLF institute in Amsterdam, and is part of the Autonomous Matter department. It is headed by Sander Tans, who is also affiliated with Delft University of Technology, and the Kavli institute of Nanoscience.
We opened new positions in December 2023! Topics include Ribosome biophysics: PhD or PostDoc, and Organoid single cell dynamics: PhD, PostDoc1, PostDoc2. You can also send an e-mail to s.tans@amolf.nl.

NEWS
- Multiple PhD and PD positions are available, e.g. through an ERC Synergy Grant with researchers from Heidelberg and ETH Zürich. Contact us if you would like to know more!
- 2021 started well for Florian with a beautiful paper in Science about ribosome-driven assembly of protein complexes.
- We congratulate Mario with his paper in Nature on protein disaggregation that was published in 2020. Also check out the movie we made about it.
- Another exciting Nature publication from our lab appeared in 2020 about how motility can drive bacterial coexistence. Here is a link to the press release.
Proteins
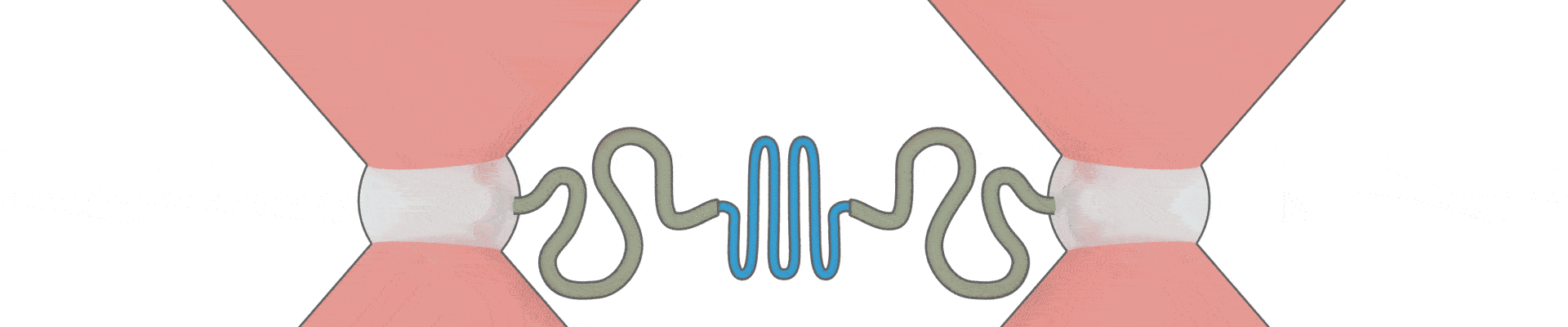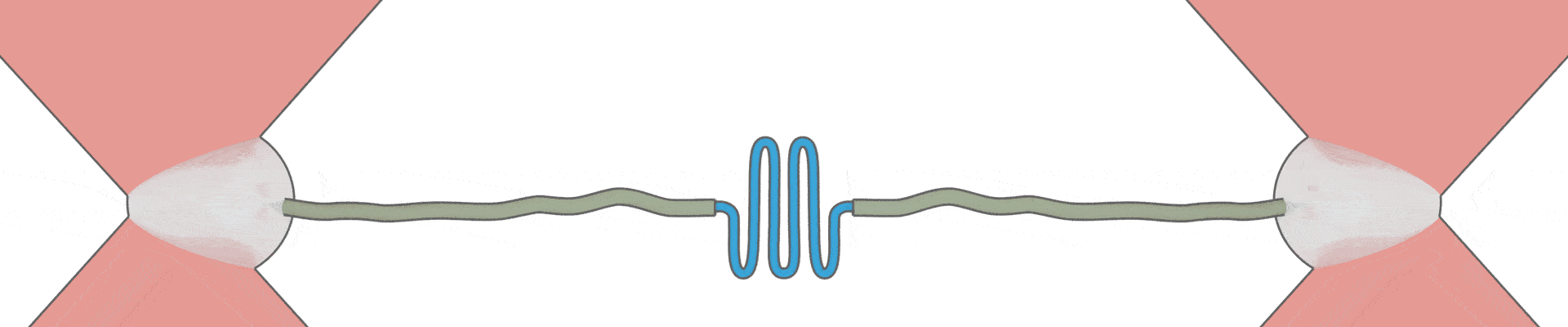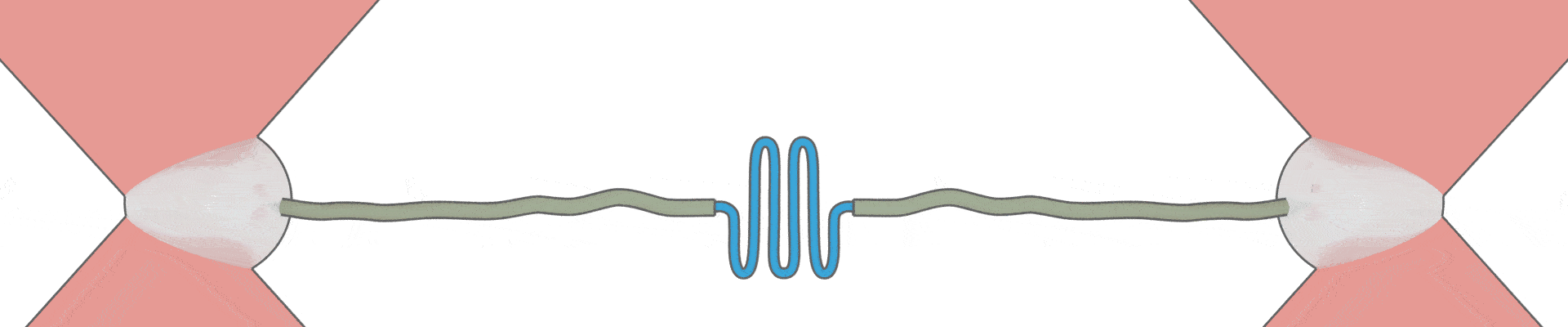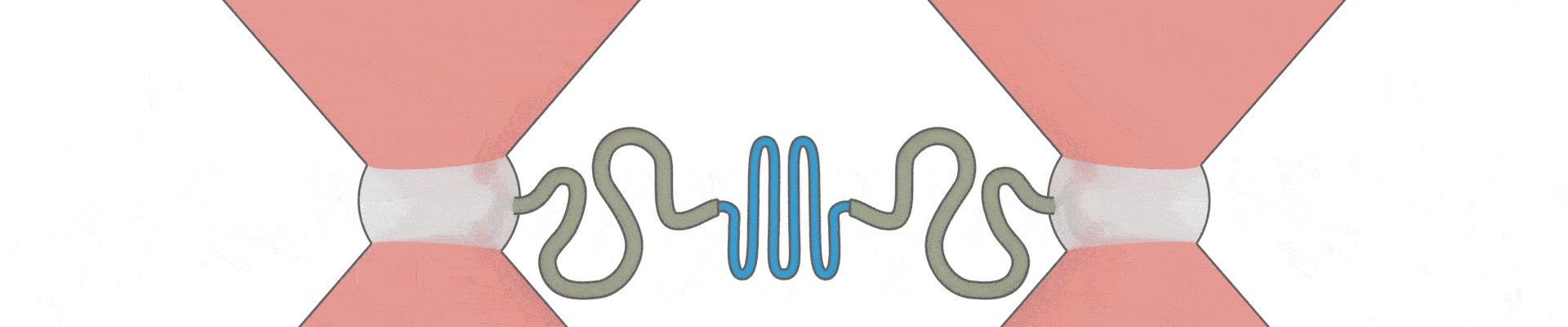
How can one protein fold another?
This intriguing question underlies all biological activity, and numerous ageing-related diseases. Recent technical breakthroughs are now making it possible to address it, by directly following the movements and forces of single protein chains within a chaperone complex.
We have pioneered the use of optical tweezers to study these dynamics of individual protein-chaperone complexes, and are currently exploiting a new range of possibilities afforded by simultaneous single-molecule fluorescence.
Key Publications
- Protein chain collapse modulation and folding stimulation by GroEL-ES (Science Advances, 2022)
- Processive extrusion of polypeptide loops by a Hsp100 disaggregase (Nature, 2021)
- Alternative modes of client binding enable functional plasticity of Hsp70 (Nature, 2016)
- Small heat shock proteins sequester misfolding proteins in near-native conformation for cellular protection and efficient refolding (Nature Communications, 2016)
- Reshaping of the conformational search of a protein by the chaperone trigger factor (Nature, 2013)
- Direct Observation of Chaperone-Induced Changes in a Protein Folding Pathway (Science, 2007)
Organoids
How do tissues self-organize?
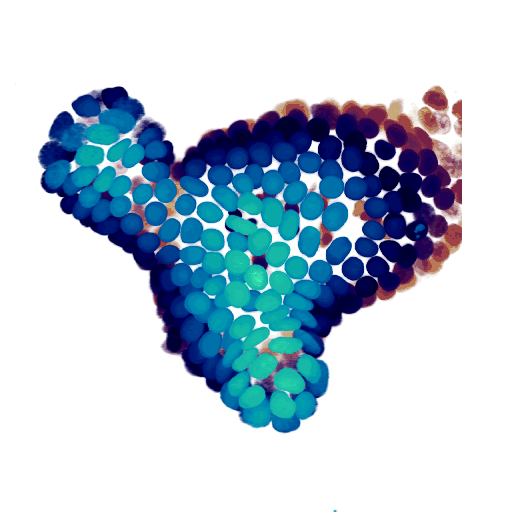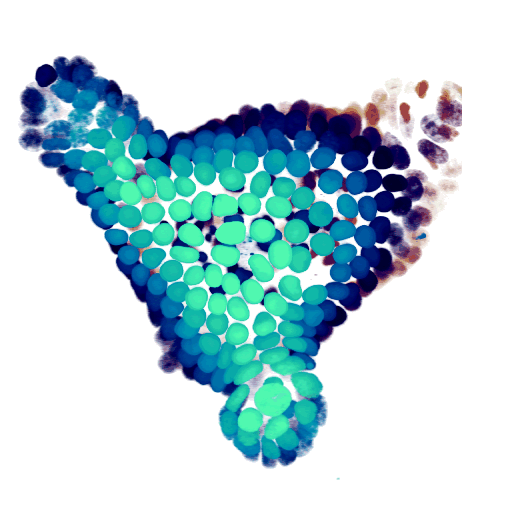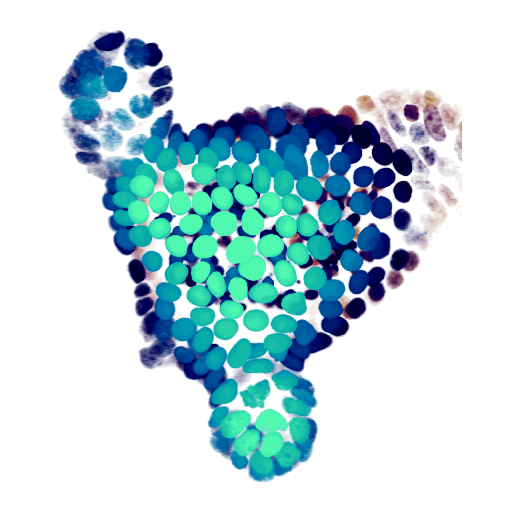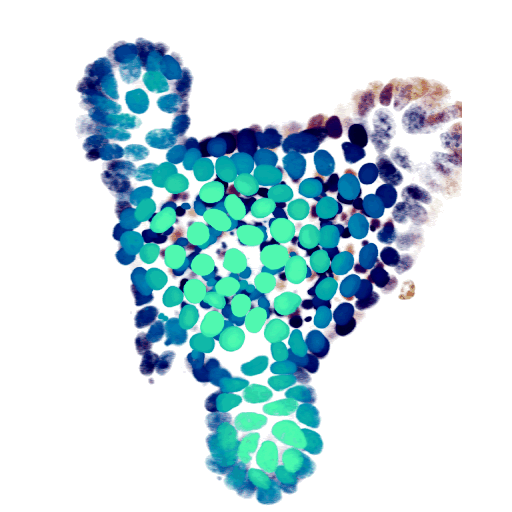
At the cellular level, we use time-lapse microscopy and image analysis to understand how multi-cellular systems self-organise, with a specific recent interest in organoids. We have developed AI-driven cell tracking and cell type inference approaches, and use these to mainly study the intestinal epithelium, often in collaboration with other groups. All our organoid work is done together with my colleague Jeroen van Zon, who runs his group also here at the AMOLF institute.
Key Publications
- Organoid cell fate dynamics in space and time (Science Advances, 2023)
- Mother cells control daughter cell proliferation in intestinal organoids to minimize proliferation fluctuations (eLife, 2022)
- ppGpp is a bacterial cell size regulator (Current Biology, 2022)
- OrganoidTracker: Efficient cell tracking using machine learning and manual error correction (PLOS ONE, 2020)
- Bacterial coexistence driven by motility and spatial competition (Nature, 2020)
- Stochasticity of metabolism and growth at the single-cell level (Nature, 2014)
Join us
Our open positions are available at the institute website. You can also contact us.